Nipype Hands-On#
Setup your analysis (import packages, set paths, etc.)#
# import all the packages that we need
from nipype import Node
from nipype.interfaces.spm import Smooth
# set general paths (we need to tell nipype where it should store the output and where the input image is)
base_dir = '/cache'
anatomical_image = '/data/single_files/anatomical.nii'
Neuroimaging example#
# Let's smooth a structural image
smooth = Node(Smooth(fwhm=[4,4,4],in_files=anatomical_image),name='smooth',base_dir=base_dir)
smooth.run()
Let’s check what happend behind the scenes#
# let's print what's inside our cache directory
!ls -la /cache/smooth
# let's check what's inside pyscript_smooth.m
!cat /cache/smooth/pyscript_smooth.m | (head --lines 20 && tail --lines 20)
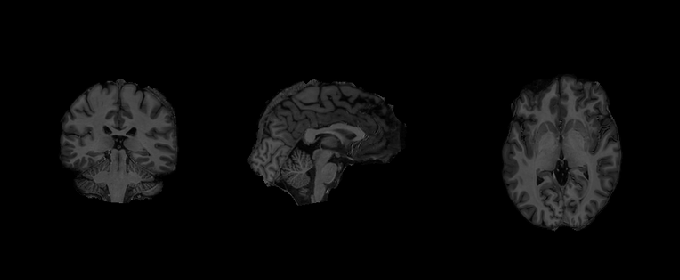
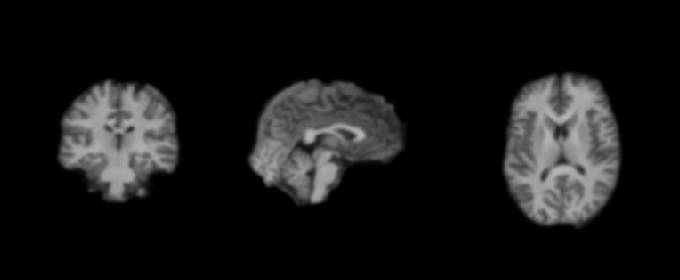
# Let's plot the image before and after smoothing
from nilearn.plotting import plot_anat
plot_anat(anatomical_image,annotate=False,draw_cross=False)
plot_anat('/cache/smooth/sanatomical.nii',annotate=False,draw_cross=False)
<nilearn.plotting.displays._slicers.OrthoSlicer at 0x7fb00d5eaf80>